Ever wanted what’s the best way to learn a new language ? well my secret is in using things I already love like R for Example . I make comparisons to familiar languages that I start to learn . It works like a charm all the time
We are going to work a lot with the dplyrpackage. You need to load the package just once when you open your environment. If you have any question, please check our other material to start learning R. However, the best way to learn is try different approaches (;
Working directory
Packages
#Working directory path
install.packages("tidyverse")
#Load library
library(tidyverse)
Help
#Help with mutate function from dplyr package
?mutate()
Load Data
CSV Files
library(readr)
#Use read_csv2
df_data <- read_csv2("data.csv")
#Using read.table function with comma separator
df_data <- read.table("my_data.csv",
sep = ",",
header = TRUE)
Excel Files
Stata Files (.dta)
Explore Data
Remember the difference between R and Stata. While in R you need to specify which object (or data) you want to work with, in Stata, the variables are loaded, so it’s easier to work with them.
/* Provides the structure of the dataset */
describe
/*Basic descriptive statistics */
summarize variable1 variable2
/*Lists the variables in the dataset */
ds
/* First 10 observations */
list in 1/10
/* Last 10 observations */
list in -1/10
/* Show first 10 observations of the */
/* first three variables of our data */
list var1-var3 in 1/10
/*View data */
browse #Provides the structure of our data
str(df_data)
#Basic descriptive statistics
summary(df_data)
#Lists the variables of our data
names(df_data)
#Show first 10 observations of our data
head(df_data, n = 10)
#Show last 10 observations of our data
tail(df_data, n = 10)
#Show first 10 observations of the
# first three variables of our data
df_data[1:10, 1:3]
# With dplyr, this would be other option
library(dplyr)
df_data %>% slice(1:10) %>% select(1:3)
#View data
View(df_data)
Missing Data
Rename variables
/* How to rename */
rename oldname newname
rename lastname lastname2
rename firstname firstname2
rename studentstatus studentstatus2
rename averagescoregrade avgscore
Label variables
label variable w "Weight"
label variable y "Output"
label variable x1 "Predictor 1"
label variable x2 "Predictor 2"
label variable age "Age"
label variable sex "Gender"
Value labels
/* Value labels for the codes */
label define label1 1 "Option 1"
2 "Option 2" 3 "Option 3" 4 "Option 4"
5 "Option 5"
/* Assign the value labels to the codes */
label values code label1#Load labelled package
library(labelled)
#Define the value labels for the codes
labels <- c("Option 1", "Option 2",
"Option 3", "Option 4",
"Option 5")
df_data$code <- labelled(df_data$code,
labels = labels)
#Define the value labels for the codes
labels <- c("Option 1", "Option 2",
"Option 3", "Option 4",
"Option 5")
df_data$code <- factor(df_data$code,
levels = 1:5,
labels = labels)In this approach, you can assign values to numeric or character variables
Group variables
Merge two datasets
/* Exact match on the key variable(s). */
use dataset1.dta, clear
merge 1:1 id using dataset2.dta
/* Each observation in the first dataset */
merge 1:m id using dataset2.dta
/* Each observation in the second dataset */
merge m:1 id using dataset2.dta
/* Each observation many-to-many merge */
merge m:1 id using dataset2.dtaRemember that we need to have our data1 in the same path
#Load dplyr package
library(dplyr)
#1:1 using inner join
df_merged_inner <- inner_join(df_1, df_1,
by = "id1")
#1:m using left join
df_merged_left <- left_join(df_1, df_1,
by = "id1")
#m:1 using right join
df_merged_right <- right_join(df_1, df_1,
by = "id1")
#m:m using full join
df_merged_full <- full_join(df_1, df_1,
by = c("id1",
"id2"))
#You can merge with more tha one variableCreate sequence/ids
Drop variables
Frequencies/Tabulation
/* Cross-tabulate 3 variables*/
tabulate var1 var2 var3
/* Cross-tabulate with row and
column percentages */
tabulate var1 var2, row col
Statistics
Summary
Linear Model
Logistic Model
/* Fit a logistic regression model */
logit foreign weight length
/* Print summary */
estimates table
Poisson Model
/* Fit a Poisson regression model */
poisson accidents weight length foreign
/* Print summary */
estimates table
Plots
Scatter plot
Line plot
Bar plot
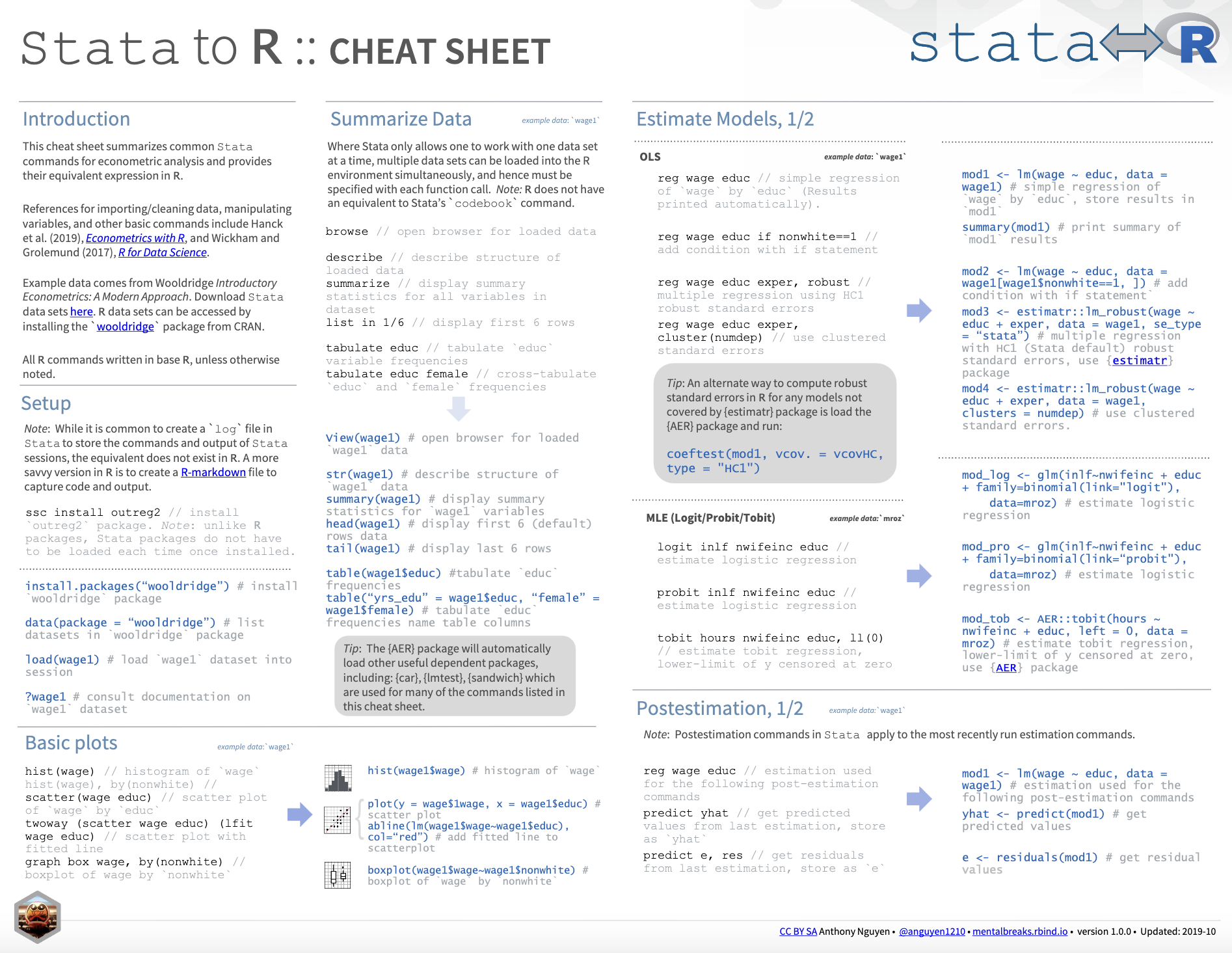
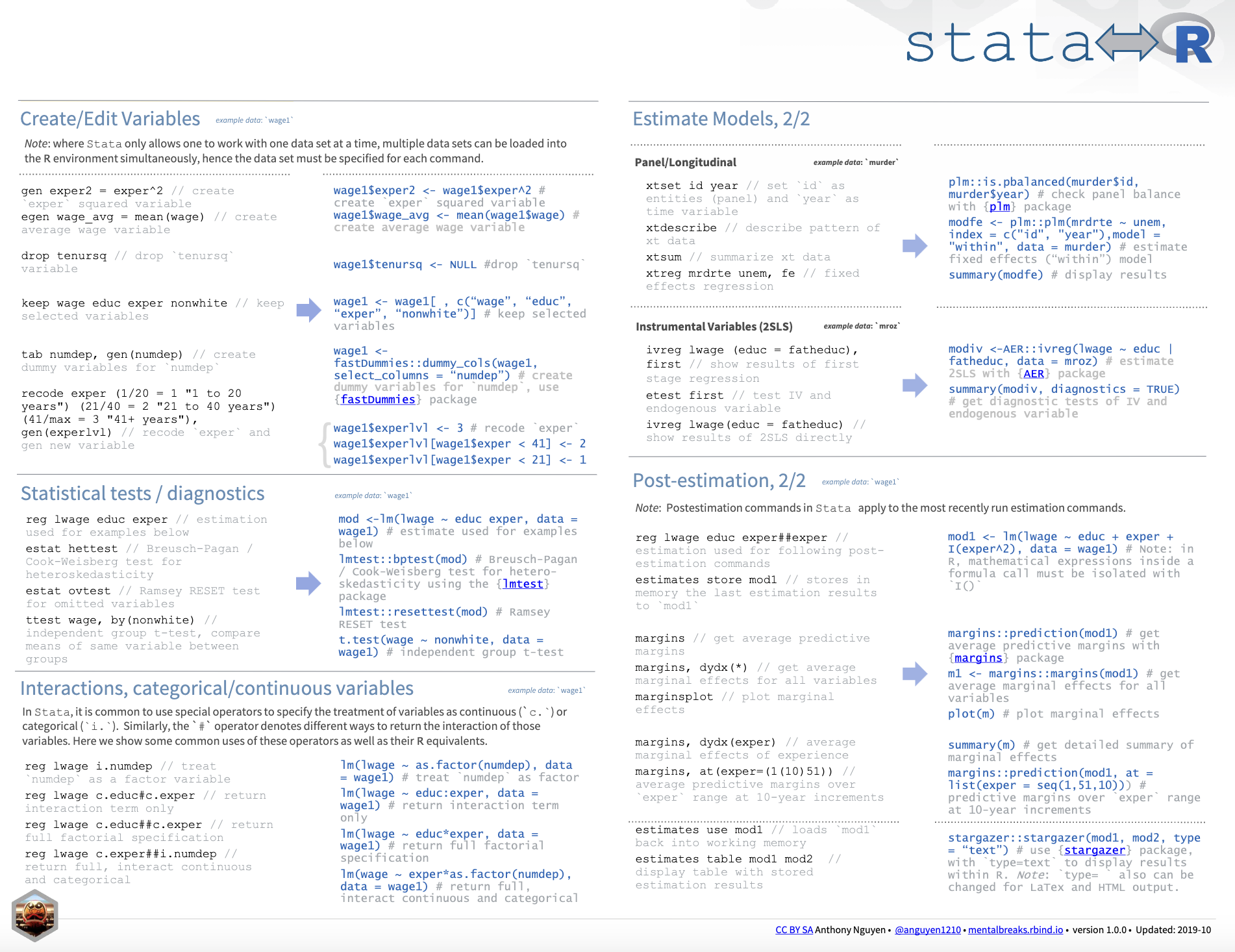
I also want to provide a good cheat sheet that contains more information about the differences between Stata and R.
Cheat Sheet: Stata to R
RStata
Believe it or not, R has a package called RStata. This package Stata interface allows the user to execute Stata commands (both inline and from a .do file) from R. I leave herethe package. Looks interesting; however, I personally prefer to use a language as expected.
Conclusion
In conclusion, the decision to transition from one to the other should be made based on the user’s specific needs and preferences. This notebook shows you the differences between Stata and R. At the end, use the language you prefer, but in my own experience, R was a life changer.
References
Jutkowitz, E., Pizzi, L. T., Shewmaker, P., Alarid‐Escudero, F., Epstein‐Lubow, G., Prioli, K. M., … & Gitlin, L. N. (2023). Cost effectiveness of non‐drug interventions that reduce nursing home admissions for people living with dementia. Alzheimer’s & Dementia.